Research Experience
What I’ve explored so far
Altered Hypothalamic Neural Activity in the mouse model of Cancer Cachexia
(Jun. 2024 - Apr. 2025)
During my undergrad studies, I took my first step into neuroscience through research on cancer cachexia—which I had witnessed in my grandfather during his battle with pancreatic cancer—a wasting syndrome marked by reduced appetite, body weight loss, and muscle atrophy. So, I first involved in establishing a murine model of cancer cachexia using Lewis lung carcinoma (LLC1) cells. During this process, mice in cachexia group showed reduced food intake and body weight loss, as well as a marked reductions in both eWAT and femoral skeletal muscle.
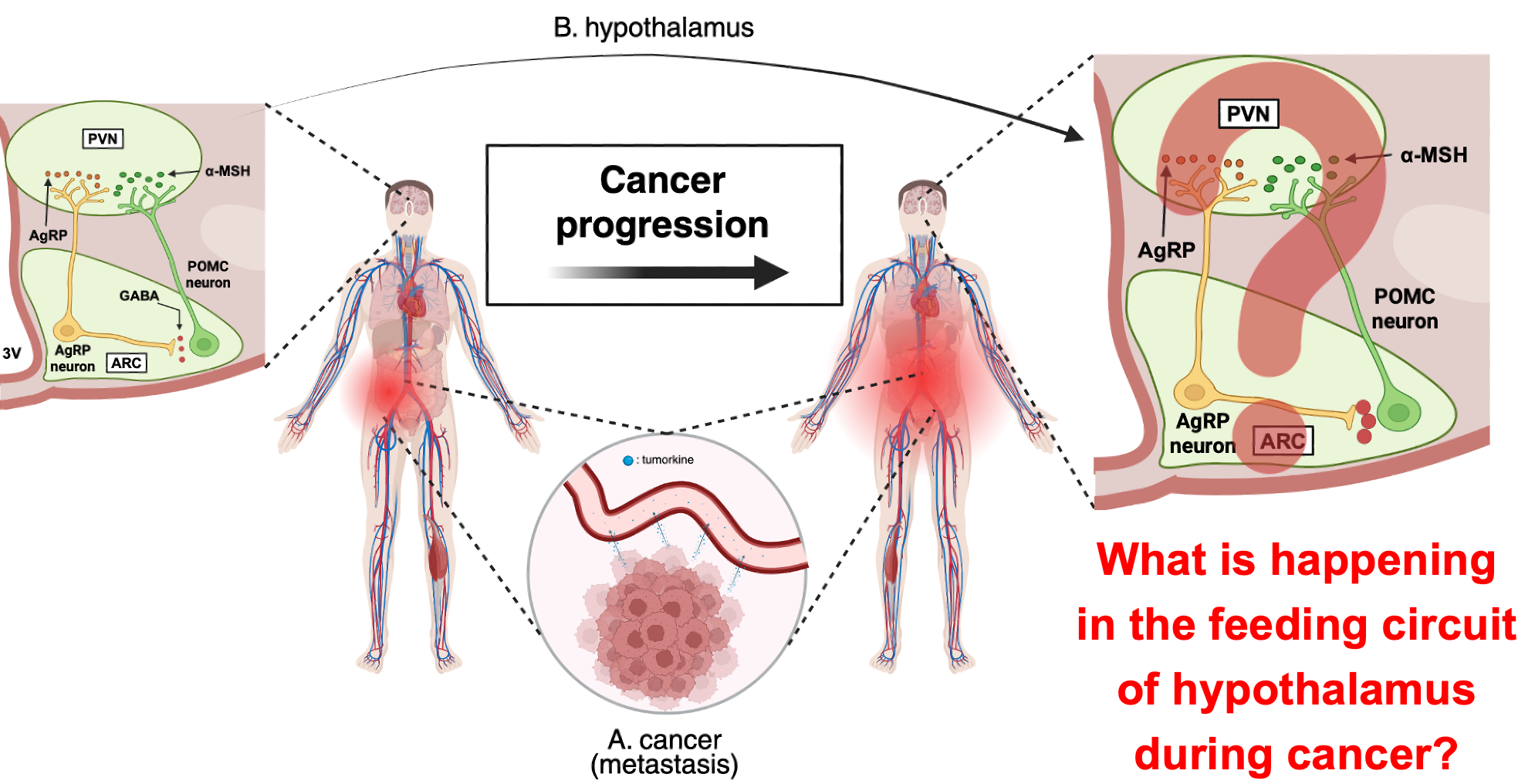
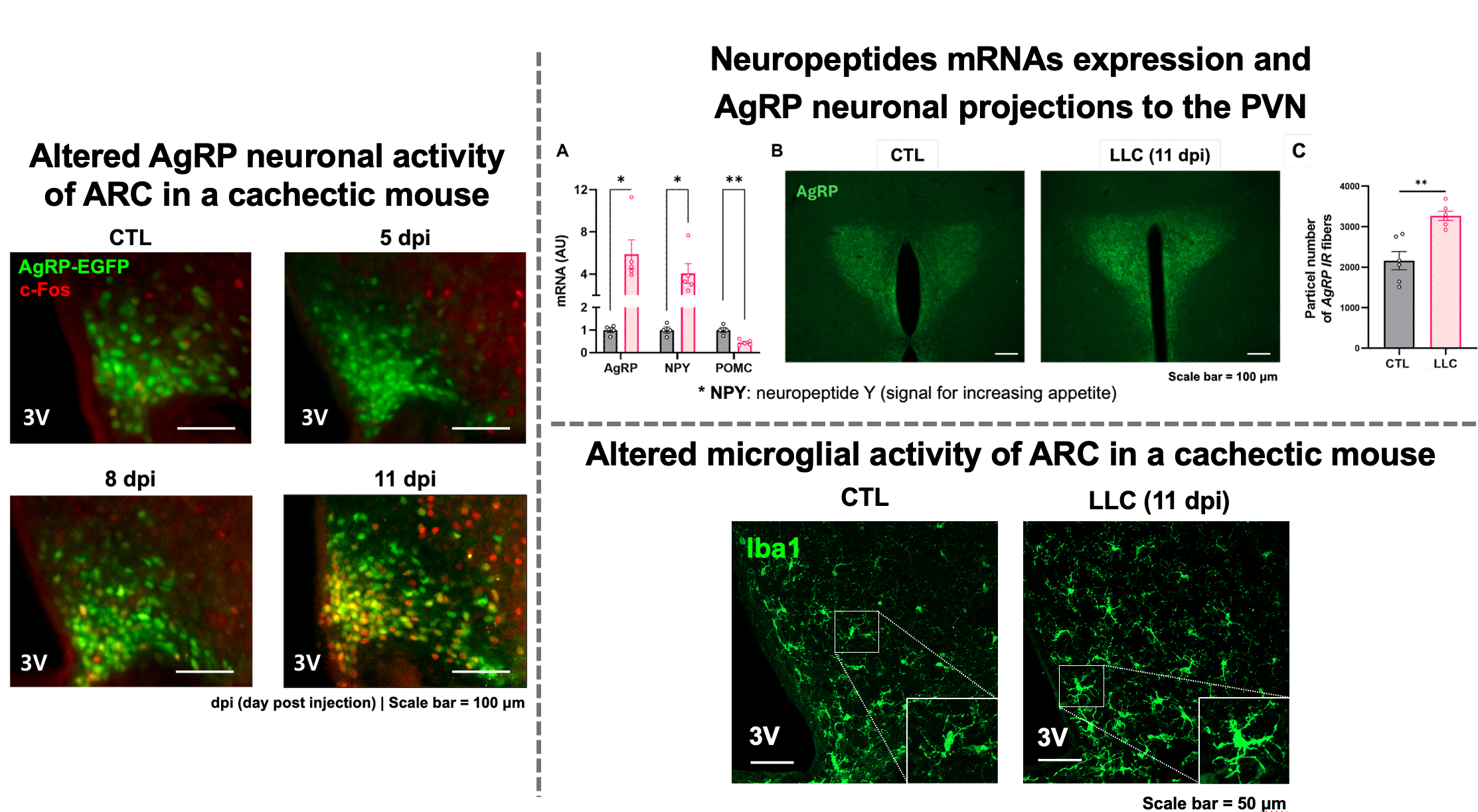
What really interested me was that, as the cancer progressed to the terminal stage, agouti-related peptide (AgRP) neurons—which normally promote hunger—became activated. Despite this activation, however, appetite did not recover. I assumed that something might have gone wrong in the downstream feeding circuit—especially within the paraventricular nucleus (PVN).
To test this, I examined the mRNA expression of neuropeptides (AgRP, NPY, and POMC) in the hypothalamus. In addition, I also examined the immunoreactivity of AgRP within the PVN. In the canonical feeding circuit, AgRP inhibit PVN neurons to promote feeding behavior. Despite strong AgRP signaling, however, PVN neurons were not inhibited, resulting in suppression of feeding behavior. Based on these findings, I reasoned that peripheral inflammatory signals from cancer might have altered the state of glial cells such as microglia, which play key neuroimmune roles in the central nervous system, thereby affecting the functional roles of multiple neurons, including appetite-regulating circuits.
Building on these insights, I want to study mainly the following.
My primary interest is understanding dynamic interactions among microglia, astrocyte, neurons, and other neural components such as oligodendrocytes, exploring how their changing states drive neuroinflammation, neuronal dysfunction, and disease progression.
From Molecular Design to Process Efficiency: Enhancing Recombinant Protein Production
(Jun. 2022 - Feb. 2023)
As an undergraduate RA at KIOST, I involved in developing a recombinant protein expression system to improve production efficiency of human EGF (hEGF). I designed gene constructs that fused a Bacillus subtilis–derived signal peptide(SP) with the E. coli–derived Trigger Factor (TF) and conducted expression and secretion assays. Through this work, I gained hands-on experience in more effective secretion of recombinant proteins. These results laid the groundwork for subsequent efforts and were later incorporated into a national R&D project launched in 2025: “Development of an Advanced Platform for Recombinant Biopharmaceutical Production Based on Marine Biological Nanomachinery.” These experiences convinced me that interdisciplinary research can be a pivotal driver in solving complex challenges in the life sciences. Moreover, I recognized that addressing real health issues ultimately requires a fundamental systems-level understanding of the human organism.
During my internship, I worked on a project to discover new broad-spectrum antibiotics using marine microbial resources. I involved in large-scale screening of over 700 isolates to identify strains capable of inhibiting superbugs, including carbapenem, ESBL, and tetracycline resistance. First, I participated in isolating marine microbes by colony picking methods and classifying these strains through 16S rRNA-based identification. I then evaluated their anti-microbial efficacy through paper disk diffusion assays. This process allowed us to obtain multiple marine microbial strains with strong antimicrobial activity.
It was a hands-on experience that helped me build a foundation in microbial screening, antibiotic efficacy testing, and translational research combating antibiotic resistance through marine-derived therapeutics.
I led a field-to-lab project investigating whether tropical disease–carrying mosquitoes were entering Jeju Island as the climate warmed. Just as a note, this was when I was a high-schooler. Using bug traps at five sites in Jeju island, we collected and morphologically identified specimens under a stereomicroscope, confirming Aedes albopictus as the dominant species. I extracted nucleic acids, performed PCR, and verified bands via agarose gel electrophoresis before sequencing. BLAST and phylogenetic analysis revealed >99% sequence match between Jeju and Vietnamese A. albopictus, strongly suggesting influx from tropical regions.
This work demonstrated how climate change can reshape disease risk landscapes, integrating ecological survey and molecular analysis to assess dengue vector introduction and persistence on the island.
On Jeju’s shores, I collected seawater, seaweed, and other tide-pool samples and cultured microbes on Marine agar/broth. I set two fish pathogens as targets—Photobacterium damselae subsp. piscicida and Edwardsiella tarda. Using a colony picking method, I screened >700 colonies and found six isolates with anti-microbial activity; two produced strong inhibition against both pathogens. I extracted genomic DNA, amplified 16S rRNA by PCR, confirmed bands by agarose gel electrophoresis, and sent amplicons for sequencing. As a result, we identified active strains as Vibrio alginolyticus and several Pseudoalteromonas spp. (piscicida, luteoviolacea, rubra, peptidolytica, elyakovii). The work provided me with end-to-end experience in sampling, microbiology, molecular identification, and interpreting experimental results for practical use.
Development of Marine-Derived Broad-Spectrum Antibiotics in response to the Superbugs Pandemic
(Aug. 2020 - Sep. 2022)
Study on the Introduction of Tropical Infectious Disease Vector Mosquitoes to Jeju Island
(Jul. 2017 - Dec. 2017)
Taxonomic Analysis of Marine Microorganisms Producing Antimicrobial Compounds
(Jun. 2017 - Nov. 2017)